Cells are the structural and functional units of all living organisms. Man is a multicellular organism, contains at least 1,014 cells. These cells differ considerably in shape, structure and function as a result of specialization. An aggregation of cells those are similar in origin, structure and function forms the tissue. Most of the metabolic activities occur at cellular level. Hence, it is essential, first to understand the basic organization of cell and functions of its components.
A typical cell, as seen by the light microscope is illustrated in Figure 1.1. It contains two compartments inner nucleus and outer cytoplasm. Nucleus contains nucleoplasm suspended with genetic material. Nuclear envelope separates nucleus from cytoplasm. Cytoplasm composed of aqueous cytosol, suspended with particles and membrane bound organelles. Externally cytoplasm is limited by plasma membrane.
ULTRASTRUCTURE
Normal cell ranges between 10 and 30 µm in diameter. The ultrastructure or finer details of typical cell, which has been revealed by the electron microscope is shown in Figure 1.2.
PLASMA MEMBRANE
The cell membrane, which completely envelops the cell, is a thin (75–100 Å), living, dynamic and selectively permeable membrane. Plasma membrane consists of specialized surface structures for attachment and for communication. These are:
- Tight junctions produce seal between adjacent cells.
- Gap junctions allow ions and electric current between adjacent cells. They may also include certain modifications to carry out physiological functions such as microvilli for absorption, invagination or infolding to carry out transportation, etc.
Biological Membranes
All biological membranes including the plasma membrane and internal membranes, which form the subcellular structures such as endoplasmic reticulum, mitochondria, lysosomes, nuclear envelope, peroxisomes, Golgi complex, etc. are similar in structure, lipoprotein in nature, consists lipids (60–40%), proteins (40–60%) and carbohydrates (1–10%). The membranes separate the cell from 2external environment and separates different parts of the cell from one another so that cellular activities are compartmentalized.
Figure 1.2: Ultrastructure of typical cell showing all cell organelles as seen in the electron microscope
ENDOPLASMIC RETICULUM
Cytoplasm is traversed by extensive network of interconnecting membrane-bound channels or cisternae (diameter of 40–50 µm), vesicles (diameter 25–500 µm) and tubules (diameter 50–190 µm) form endoplasmic reticulum (ER) (Fig. 1.3).
Membranes of ER are continuous with plasma membrane and outer nuclear envelope. There are two basic morphological types:
- Rough endoplasmic reticulum (RER) possesses rough surface due the attachment of ribosomes. RER occurs mainly in the form of cisternae and concerned with protein synthesis.
Endoplasmic reticulum provides skeletal framework to the cells and gives mechanical support to the colloidal cytoplasm. It also plays a role in detoxifying the xenobiotics.
GOLGI COMPLEX
- Golgi complex is membrane-bound structure similar to ER, discovered in 1873 by Camillo Golgi.
- It is a stack of flattened membrane vesicles (cisternae) surrounded by network of tubules of 300–500 Å diameter. Cisternae are gently curved, convex part (cis side) faces ER and concave part (trans side) locates near the plasma membrane (Fig. 1.4).
- Golgi complex functions in association with ER, is a center of reception, finishing, packaging and transportation of variety of materials.
- Proteins synthesized in ER is added with sulfate, carbohydrates, lipid moieties, etc. and dispatched in the form of secretory vesicles.
- Golgi complex also gives rise to lipoprotein of plasma membrane and lysosomes.
LYSOSOMES
- Lysosomes are packets of hydrolases.
- These are spherical 1 µm in diameter surrounded by tough carbohydrate-rich lipoprotein membrane enclosing about 50 types of hydrolases such as proteases, lipases, carbohydrases, nucleases, transferases, sulfatases, etc.
- Lysosomes provide an intracellular digestive system through which macromolecules, foreign bodies, worn out and unwanted structures are got digested.
PEROXISOMES
- Peroxisomes are circular membrane bound organelle having about 0.25 µm diameters contain enzymes peroxidases and catalase.
- Peroxisomes detoxify various toxic substances and metabolites through peroxidative reactions catalyzed by peroxidases. Catalase degrades hydrogen peroxide (H2O2) resulted from the breakdown of fatty acid and amino acids.
MITOCHONDRIA
- Mitochondria are spherical, oval or rod-like bodies. It is about 0.5–1 µm in diameter and up to 7 µm in length (Fig. 1.5).
- Mitochondria are considered to be the powerhouse of the cell, where energy released from oxidation of foodstuffs is trapped as chemical energy in the form of adenosine triphosphate (ATP).
- Mitochondria are respiratory center of cell where pyruvate oxidation, citric acid cycle, electron transport chain and ATP generation takes place. The β-oxidation of fatty acid and ketone body synthesis also takes place.
CENTRIOLES
- Two cylindrical rods-shaped structures of 0.3–0.7 µm lengths and 0.1–0.25 µm diameters, which lie at right angles to one another near the nucleus is called centrioles.
- Centriole is an array of 9-triplet microtubules equally spaced from central axis, made up of structural protein tubulin.
- Centrioles form mitotic poles during cell division.
- They also give rise to cilia and tail of sperm.
NUCLEUS
- Nucleus is a cell center, a prominent spherical structure where genetic material is confined.
- All cells in the human body contain nucleus, except matured red blood cells (RBCs) and upper dead skin cells.
- Generally nucleus is spherical or oval in shape and of 3–25 µm in diameter. But squamous epithelial cells contain diskoidal and it is multilobed in polymorphonuclear leukocytes.
- Nuclear envelope, which encircles the nucleus, consists of outer and inner nuclear membranes, typical lipoprotein membranes.
- Outer nuclear membrane is continuous with membranes of the ER and is found attached with ribosomes on its outer surface.
- Nuclear envelope contains numerous nuclear pores of 100–1,000 Å diameter, which regulates the nucleocytoplasmic trafficking of ions, nucleotides, proteins, messenger RNA (mRNA), transfer RNA (tRNA) and ribosomal subunits.
- Nucleoplasm consists of genetic material (chromosomes) and nucleolus.
- Nucleolus is a ribonucleoprotein structure and is the site of formation of ribosomal subunits.
- Nucleoplasm is composed of mainly the nucleoproteins, proteins, enzymes, minerals, organic and inorganic substances.
TRANSPORT ACROSS THE MEMBRANE
Biological membranes are lipoprotein viscous barriers, exist around all the living cells and also form structural and functional component of all the cell organelles. Membranes contain mainly lipids, proteins and very little amount of carbohydrates. The contents of these vary according to the nature of the membrane. Lipids are mainly amphipathic phospholipids, glycolipids and cholesterol. Proteins are of two types:
- Peripheral or extrinsic proteins: Loosely held to the surface of the membrane and can be easily separated, e.g. cytochrome c of mitochondria.
Organization of biological membranes, the arrangement of lipids and proteins was best explained in fluid mosaic model of Singer and Nicolson (1972) (Fig. 1.6). According to this model, membrane is a viscous fluid phospholipid bilayer in which globular proteins are inserted in a mosaic pattern. Amphipathic phospholipid consists of a polar phosphate head, a glycerol neck and two non-polar fatty acid tails. Hydrophobic tails or fatty acids form the middle core of lipid bilayer and hydrophilic heads line both the sides. Both phospholipids and proteins are amphipathic in nature, and form a permeability barrier. Degree of saturation and unsaturation of fatty acids, presence of cholesterol and carbohydrates regulate the fluidity and movement of molecules. Hydrophilic heads of inner and outer surface keep constant circulation of water. But hydrophobic fatty acid core acts as selective permeable barrier, saves the cells and cell organelles from osmotic shocks. Important function of the membrane is to withhold unwanted molecules, but permit entry of molecules necessary for cellular metabolism.
Transport across the membrane occurs in following ways:
- Passive transport
- Active transport
- Exocytosis
- Endocytosis.
Passive Transport
Passive transport of molecules across the membrane is along the concentration gradient without using energy. Movement of molecules from higher concentration to lower concentration takes place without using energy. Solutes and gases enter into the cells passively. They are driven by the concentration gradient. The rate of transport is directly proportional to the concentration gradient of that solute across the membrane. Passive transport of molecules across the biomembranes is in two ways.
Simple Diffusion
Small uncharged molecules such as water (H2O), oxygen (O2), carbon dioxide (CO2), methane (CH4), other gases, urea, ethanol, etc. cross the lipid bilayer by simple diffusion.
Facilitated Diffusion or Carrier-mediated Passive Transport
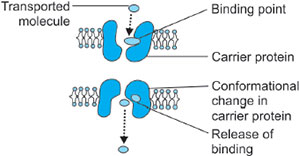
Diffusion of molecules across the membrane along the concentration gradient occurs through carrier proteins or permeases. It differs from simple diffusion in certain aspects. Firstly, the process is stereospecific, i.e. only one of the two possible isomers, L and D, is transported. Secondly, it shows saturation kinetics. Thirdly, a carrier is required for transport across the membranes.
Mechanism: The carrier proteins or permeases are specific integral membrane proteins and are highly specific for molecules, which they transport. Carrier proteins are specific for individual sugars, amino acids, phosphate, etc. 6Whenever there is a concentration gradient of a solute across the membrane, solute molecules from hypertonic side bind to specific permease of the membrane. This binding triggers some conformational change producing a pore or tunnel in the carrier protein through which ions, glucose, etc. may cross. After the transportation permease regains its original structure (Fig. 1.7).
Active Transport
Active transport of molecules across the membrane occurs against the concentration gradient using energy. Molecules are transported from lower concentration (hypotonic) to higher concentration (hypertonic) with the use of energy (Fig. 1.8).
In all cells, a significant portion of energy goes in maintaining the concentration gradient of ions across plasma membrane and intracellular membranes.
In human RBC, 50% of (cellular metabolism) energy is used for the above purpose. Active transport is of two types:
- The ATP-driven active transport or primary active transport:
- Transmembrane proteins or carrier proteins form channels to bring the transport of molecules and ions across biological membranes using energy from ATP.
- The most important active transport in the cells is sodium-potassium adenosine triphosphatase (Na+/K+-ATPase) pump.
- All cells maintain high internal concentration of K+ and low concentration of Na+.
- This Na+/K+ gradient across the membrane is maintained using energy from hydrolysis of ATP.
- The ATPase is a large carrier protein, hydrolysis of ATP brings the binding of 3 Na+ to ATPase, which carries some conformational changes in ATPase, so that 3 Na+ is pumped outside and in exchange 2 K+ is pumped in opposite direction (refer Fig. 1.8).
- Ion-driven active transport or secondary active transport:
- Secondary active transport takes place in the presence of ionic gradient maintained across the membrane by primary active transport.
- For example, glucose absorption in intestinal epithelial cells. The concentration gradient maintained by Na+/K+-ATPase pump across the cell brings the symport of Na+ and glucose molecules into the cell.
EXOCYTOSIS AND ENDOCYTOSIS
Exocytosis
Secretions of cell such as proteins, lipids and carbohydrates are released out of the cell through exocytosis. These secretions are packed in the form of secretory vesicles. 7As per necessary stimulation these vesicles move towards the plasma membrane and fuse with the plasma membrane. In this way materials inside the vesicles are externalized. For example, release of acetylcholine from synaptic vesicles in presynaptic cholinergic nerves; release of trypsinogen by pancreatic cells; release of insulin by beta cells of Langerhans, etc.
Endocytosis
Endocytosis is the mechanism by which cells uptake macromolecules in the form of endocytic vesicles. Plasma membrane invaginates and encloses the materials, which results into vesicles. There are two types, phagocytosis and pinocytosis (Fig. 1.9).
Phagocytosis
Phagocytosis is the ingestion of large particles such as bacteria, cell debris, etc. Plasma membrane invaginates in the form pseudopodia and encloses the particles in the form of phagosome. Materials of phagosomes will be digested by lysosomes. For example, engulfment of bacteria by macrophages and granulocytes.
Pinocytosis
Pinocytosis is the uptake of nonspecific or specific extracellular molecules in the form of endocytic vesicles. Later it is termed as receptor-mediated endocytosis. Plasma membranes internalize these receptor-attached molecules in the form of vesicles. For example, uptake of chylomicrons by liver cells; internalization of low-density lipoprotein (LDL) through LDL receptors of plasma membrane.
TRANSPORT SYSTEMS
The transport systems may be divided into three categories (Fig. 1.10).
Uniport System
Carrier proteins, which simply transport a single solute from one side of the membrane to the other, are called uniports, e.g. transport of glucose through the membrane.
Symport and Antiport System
Transport of one solute depends on the simultaneous transfer a second solute, either in the same direction (symport) or in the opposite direction (antiport). Both symport and antiport are collectively called cotransport system.
Symport: Transport of glucose and Na+ into the intestinal epithelial cell from the gut.
8Antiport: For example, Na+/K+-ATPase pump, chlorine (Cl−) and bicarbonate (HCO3−) is exchanged in erythrocytes.
CELL FRACTIONATION
The study of biochemical properties of individual organelles requires subcellular fractionation. The subcellular fractionation involves breaking of cell by means of mechanical force to purify organelles. The steps involved in are:
- Mince the tissue using buffer.
- Tissue is carefully broken up in homogenizer using isotonic 0.25 M sucrose solution (the sucrose solution is preferred because is not metabolized, does not pass through the membranes readily and does not cause interorganelles to swell).
- The gentle homogenization in an isotonic sucrose solution ruptures the cell membrane and keeps most of the organelles intact. But ER is broken into small pieces that form microsomes.
- Homogenate is drained to remove connective tissue and fragments of blood vessels by stainless steel sieve.
- The homogenate, thus obtained, is centrifuged at a series of increasing centrifugal force.
The nuclei and mitochondria differ in size and specific gravity, and therefore sediment at different rates in a centrifugal field and can be isolated from the homogenate by differential centrifugation (Fig. 1.11). The dense nuclei are sedimented first, followed by the mitochondria and finally microsomal fraction. The soluble remnant is the cytosolic portion.
The mitochondria isolated in this way are contaminated with lysosome and peroxisomes. These may be separated by isopycnic centrifugation technique (Fig. 1.12). In this technique, a density gradient is set up in a centrifuge tube (the density of the solution in the tube increases from top to the bottom). Sucrose is used as medium and colloidal materials such as pecroll, which form density gradients with a low-osmotic pressure, are often used. Particles are sedimented to an equilibrium position at which their density equals that of the medium at that point in the tube. Different organelles are separated according to their density.
9The purity of the isolated subcellular fraction is assessed by analysis of marker enzymes, which are located exclusively in a particular fraction and are specific to that fraction (Table 1.1). Analysis of marker enzymes confirms the degree of purity and contamination.
1. Briefly discuss the ultrastructure of a typical cell.
2. Add a note on the structural aspects of mitochondria and mention the metabolism, which takes place in mitochondria.
3. Explain the fluid mosaic model of plasma membrane.
4. Write the features and importance of active transport mechanism.
5. How do you explain the ATP-driven active transport and ion-driven active transport?
6. Mention few significances of endocytosis and exocytosis.
7. What is ion-driven active transport? Explain with an example.
8. Explain uniport and antiport transport mechanism with an example.
9. Why do we call mitochondrion as a powerhouse of the cell?
10MULTIPLE CHOICE QUESTIONS
1. Concerning plasma membrane, one of the following statements is not true:
- Plasma membrane consists of specialized surface structures for attachment and for communication
- Tight junctions produce seal between adjacent cells
- Gap junctions do allow ions and electric current between adjacent cells
- Consists of proteins, lipids and carbohydrates
2. Cytoplasm is traversed by extensive network of interconnecting membrane-bound channels or cisternae, vesicles and tubules, which form:
- Endoplasmic reticulum
- Golgi complex
- Ribosomes
- Microsomes
3. Concerning the Golgi complex, all of the following statements are true except:
- It is a membrane-bound structure
- It is a stack of flattened membrane vesicles
- It does not give rise to lipoprotein of plasma membrane
- It helps in packaging and transportation of variety of materials
4. Concerning mitochondria, one of the following statements is incorrect:
- It is considered to be the powerhouse of the cell
- They are respiratory center of cell where pyruvate oxidation takes place
- It accommodates for glycolysis
- It has electron transport chain
5. Nucleus:
- Present in all cells of the body
- Does not have nuclear envelope
- Absent in RBCs
- Exists in different shapes
6. Concerning passive transport, one of the following statements is incorrect:
- It requires ATP
- It requires carrier protein
- Occurs along the concentration gradient
- Process is stereospecific
7. Concerning active transport, one of the following statements is incorrect:
- Transport of molecules across the membrane is against the concentration gradient
- It is energy dependent
- Most important active transport in cells is Na+/K+-ATPase pump
- 2 Na+ pumped outside and in exchange 3 K+ pumped in opposite direction
8. Glucose absorption in intestinal epithelial cells is:
- Ion-driven active transport
- Facilitated diffusion
- Passive transport
- Does not depend on concentration gradient
9. Transport of macromolecules takes place through the following mechanisms except:
- Diffusion
- Phagocytosis
- Pinocytosis
- Exocytosis
10. All of the following are the examples for endocytosis except:
- Uptake of chylomicrons by liver cells
- Internalization of LDL through LDL receptors of plasma membrane
- Uptake of glucose by intestinal cells
- Engulfment of bacteria by macrophages
11. The main function of mitochondria is:
- DNA synthesis
- Protein processing and packaging
- ATP production
- RNA synthesis
- DNA synthesis
- Protein processing and packaging
- ATP synthesis
- RNA synthesis
13. The following are true of plasma membranes except:
- They are made up of a double layer of lipid molecules in which proteins are embedded
- The lipid membranes include phospholipids and cholesterol
- The plasma membrane has RNA-binding sites on the inside surface of the membrane resembling rough endoplasmic reticulum
- The plasma membrane has both integral membrane proteins and peripheral membrane proteins
14. The function of smooth endoplasmic reticulum is:
- Protein synthesis
- Regulation of intracellular calcium distribution
- Excretion
- Maintain the skeleton of the cell
15. All of the following are the functions of lysosomes except:
- Phagocytosis
- Pinocytosis
- Exocytosis
- Breakdown of some intracellular materials
16. Hydrolytic enzymes are found in:
- Golgi apparatus
- RER
- Lysosomes
- Ribosomes
17. The site of lysosomes can be seen using a specific histochemical reaction called:
- Alkaline phosphatase
- Acid phosphatase
- Peroxidase
- Succinic dehydrogenase
18. Organelles most notable for producing and degrading hydrogen peroxide are:
- Lysosomes
- Mitochondria
- Golgi bodies
- Peroxisomes
19. The function of attached ribosomes to RER is to synthesize:
- Lipid
- Carbohydrate
- Protein that will be secreted by the cell
- Glycogen
20. Ribosomal RNA is formed in:
- The euchromatin
- The nucleolus
- The RER
- The heterochromatin
21. Glycogen can be demonstrated using:
- Best's carmine
- H and E
- Sudan black
- Silver
22. Euchromatin is predominant in:
- Present in the nuclei of metabolically active cells
- Present in the nuclei of metabolically inactive cells
- Special type of stain
- Type of cell organoids
23. The nucleolus is formed of:
- Protein and DNA
- Protein only
- Chromatin
- Protein and RNA
24. The nuclear pore:
- Is hexagonal in shape
- Is bridged by a unit membrane
- Allows for communication between the nucleus and the cytoplasm
25. The feature of phospholipids that is essential for their role in biological membranes is:
- Form strong rigid membranes
- Extremely hydrophobic
- Possess hydrophilic and hydrophobic portions
- Extremely hydrophilic
1. c | 2. a | 3. c | 4. c | 5. c |
6. a | 7. d | 8. a | 9. b | 10. c |
11. c | 12. b | 13. c | 14. b | 15. c |
16. c | 17. b | 18. d | 19. c | 20. b |
21. a | 22. a | 23. d | 24. d | 25. c |