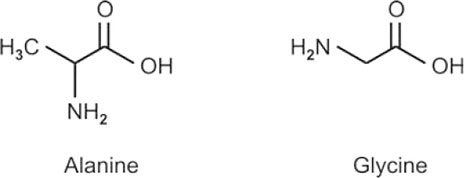- Name three aromatic amino acids. Write the structure of one of them.
The three aromatic amino acids are: phenylalanine, tyrosine, and tryptophan.
The three hydrophilic amino acids are: aspartic acid, glutamic acid, and lysine
Essential amino acids are methionine, threonine, tryptophan, valine, isoleucine, leucine, phenylalanine and lysine.
Semi essential amino acids are arginine and histidine.
The three heme proteins are: hemoglobin, myoglobin, and cytochrome oxiodase
- Hemoglobin transports oxygen from lungs to the tissues
- Myoglobin stores the oxygen in the muscle.
- Cytochrome oxidase is a component of electron transport chain and transfers electrons from cytochrome c to molecular oxygen. It also acts as a proton pump during electron transport in the respiratory chain.
peptides of biological importance.
Aspartame: It is a dipeptide made up of aspartic acid and modified phenylalanine. It is used as a sweetener. It is a non-caloric sweetener.
Glutathione is a tripeptide. It is γ-glutamyl cystenyl glycine. It protects the membrane structures of RBC from oxidative damage and helps in keeping enzymes in active state. It is required to reduce methemoglobin back to hemoglobin and transports amino acids across the membrane in the intestine and kidney.
Angiotensin II and III: Angiotensin II has eight amino acids whereas angiotensin III has seven amino acids. They cause hypertension.
Oxytocin: Oxytocin is composed of nine amino acids. It stimulates contraction of smooth muscles of uterus and also helps in ejection of milk from the mammary gland.
Vasopressin: It has nine amino acids. It is also called antidiuretic hormone (ADH), and helps in reabsorption of water from distal convoluted tubules and collecting ducts. It increases the blood pressure.
- Name sulfur containing amino acids. Write the biological functions of any one of them in the body.
Sulfur containing amino acids are methionine, and cysteine.
Biological functions of methionine are as follows:
- Methionine is an essential amino acid and glycogenic amino acid
- Methionine gives rise to cysteine
- Activated methionine (S-adenosyl methionine) is a methyl donor and participates in transmethylation reactions.
- Methionine participates in polyamine and lipoprotein synthesis.
- Methionine is required for protein biosynthesis and it is also the first amino acid to be incorporated during translation (protein biosynthesis).
- It is a dietary source of sulfur to the body.
- Write the composition of glutathione. What its role in erythrocytes.
Glutathione is γ-glutamyl cysteinyl glycine. It is made up of three amino acids: glutamic acid, cysteine, and glycine.
Glutathione functions as antioxidant (free radical scavenger) in erythrocytes: hydrogen peroxide is continuously formed inside the RBC. Hydrogen peroxide destroys biomembranes by lipid peroxidation and also oxidizes hemoglobin to methemoglobin. This leads to hemolysis and decreased life span of RBC. Damage caused by hydrogen peroxide is prevented by detoxication of hydrogen peroxide by glutathione peroxidase; a selenium-containing enzyme present in RBC that require reduced glutathione. Reduced glutathione is regenerated from oxidized glutathione with the help of glutathione reductase using NADPH produced in pentose phosphate pathway. Reduced glutathione also prevents the oxidation of -SH groups of membrane proteins and hemoglobin.
- Define isoelectric pH of amino acid and write its importance.
The pH at which the amino acids exist as zwitterions and carries no net charge is known as the isoelectric pH (pI) of amino acids. At isoelectric pH, the amino acid is electrically neutral and does not move either towards anode or cathode in an electric field.
At pI, the amino acid is least soluble and has minimum buffering capacity. Hence precipitation of an amino acid is maximal at its isoelectric pH.
- Write the structure of typical amino acid. Name any three nonprotein amino acids.
The nonprotein amino acids are those, which are not constituents of proteins. The few examples of nonprotein amino acids are ornithine, citrulline, homocysteine, and β-alanine.
- Define proteins, and enumerate four biologically important functions of proteins.
Proteins are linear polymers of L-amino acids linked by peptide bonds. Proteins are soluble, colloidal, high molecular weight (non-dialyzable) organic substances, which can undergo denaturation.
Four biologically important functions of proteins are as follows:
- Biological catalysts (enzyme function)
- Defense of the body (immunoglobulins)
- Regulation of cellular activities (hormones)
- Transport of various substances (carrier proteins)
- Classify proteins based on functions with suitable examples.
Proteins are classified as follows based on their functions.
- Biological catalysts or enzymes: e.g. salivary amylase, pepsin, and lactase.
- Structural proteins: e.g. collagen, keratin, and elastin.
- Transport proteins: e.g. hemoglobin, transferring, and serum albumin.
- Protective proteins: e.g. immunoglbulins, blood clotting factors, and interferons.
- Contractile or motile proteins: e.g. actin, myosin, and tubulin.
- Regulatory proteins: e.g. hormones like insulin, receptors like insulin receptors.
- Storage proteins: e.g. myoglobin, ferritin, and ovalbumin.
- Genetic proteins: e.g. histones, protamines, and non-histones.
- Proteins with other functions e.g. sweet proteins – thaumatin, and monellin.
- Classify amino acids based on chemical nature with suitable examples. Write the structure of one basic and one acidic amino acid.
Based on chemical nature, amino acids are classified into three types: acidic amino acids, basic amino acids, and neutral amino acids.
- Acidic amino acids are monoamino dicarboxylic acids, which are acidic in nature.Examples are aspartic acid, and glutamic acid.
- Basic amino acids are dibasic monocarboxylic acids, which are basic in nature. Examples are lysine, arginine, and histidine.
- Neutral amino acids are neutral in character and are further subdivided into different types:
- Aliphatic amino acids: These amino acids contain aliphatic side chains. Examples are glycine, alanine, valine, isoleucine, and leucine. The last three are known as branched chain amino acids because their side chains are branched.
- Aromatic amino acids: These amino acids contain aromatic side chains. Examples are phenylalanine, tyrosine, and tryptophan. Tyrosine is slightly acidic due to phenolic side chain group.
- Hydroxy amino acids: These contain hydroxyl group in the side chain. Examples are serine, and threonine.
- Sulfur containing amino acids: They contain sulfur in the side chain. Examples are methionine, and cysteine
- Amides of acidic amino acids: Side chain carboxylic groups of aspartic acid and glutamic acid are neutralized by amide formation. Examples are asparagine, and glutamine.
- Imino acid: It contains an imino group (in the ring) instead of the usual amino group present in other amino acids. Example is proline.
- Classify amino acids based on their side chains, and their importance in influencing the properties of protein.
Various side chains (R groups) of amino acids are classified into four groups based on polarity: nonpolar side chains, nonionic polar side chains, polar side chains with anionic (negatively charged) groups, and polar side chains with cationic (positively charged) groups.
Nonpolar side chains: aliphatic group of valin, and aromatic groups of phenylalanine and tryptophan are examples of this group. They contribute to the stability of tertiary structure. Non-polar amino acids do not influence the properties of proteins. Tryptophan and phenylalanine absorbs ultraviolet light energy.
Non-ionic polar side chains: Hydroxyl, sulfhydryl, and amide groups are examples of hydrophilic side chains. These groups contributing to the stability of protein structure by hydrogen bonding, but do not influence the properties of proteins. The -OH groups attract water and cause hydration of proteins. Tyrosine also absorbs ultraviolet light energy.
Polar side chains with anionic groups: Carboxylic groups are examples for this group. These groups contribute to the charged properties and amphoteric nature of proteins. On the alkaline side of isoelectric pH, the protein exists as anions (-COO−), and can be precipitated by heavy metal ions such as mercury (Hg2+) and lead (Pb2+), and also can be stained by basic dyes such as methylene blue and hematoxylin. The -COOH groups also cause hydration of protein.
Polar side chains with cationic groups: The side chain groups of arginine, lysine, and histidine that can be protonated are the examples of this 6group. These groups contribute to amphoteric nature, and charged properties of proteins. On the acidic side of isoelectirc pH the protein is protonated and exists as cation. The -+NH3 group of protein can be precipitated by alkaloidal reagents such as sulfosalicylic acid and tannic acid and also can be stained by acidic dyes like eoasin and scarlet red. The -NH2 groups attract water and cause hydration of proteins.
- What is isoelectric pH (pI) of a protein? In what ionic form do proteins circulate in blood? What is its physiological significance?
The pH at which the protein exists as zwitterion and carry no net charge is known as isoelectric pH (pI) of the protein. At isoelectric pH, the protein is electrically neutral and does not move either towards anode or cathode.
At physiological pH (pH7.4), the plasma proteins circulate in blood as anions.
Physiological significance of pI of plasma proteins is as follows:
Almost 50% of plasma calcium and zinc (Zn2+) are bound to negatively charged plasma proteins, particularly albumin and is transported in the blood. Copper (Cu2+) is also loosely bound to plasma albumin and is transported in the blood.
Negatively charged plasma proteins (proteinate ions), neutralize the metabolic acids by combining with hydrogen ions.
Plasma contains higher concentration of solutes compared to interstitial fluid because of anionic plasma proteins. This provides net osmotic gradient to maintain plasma volume.
- Write the structure of insulin. How does it differ from proinsulin? Write three biochemical functions of it.
Proinsulin differs from insulin in having an extra connecting peptide (C-Peptide). The C-peptide is 35 amino acids long and connects ‘A’ chain and ‘B’ chain in proinsulin.
Three biochemical functions of insulin:
- Insulin is an anabolic hormone and promotes synthesis of protein fat and glycogen. Insulin lowers the blood glucose level by promoting glucose utilization by the peripheral tissues and glycogenesis in liver, and also by inhibiting gluconeogenesis in liver.
- Insulin stimulates the synthesis of triacylglycerols and also decreases lipolysis by inhibiting hormone-sensitive lipase.
- Insulin increases the entry of amino acids into the cells, promotes protein synthesis and inhibits degradation of proteins.
- Write the structure of peptide bond. How is it formed? How does it influence the secondary structure of protein?
The bonds connecting the α-carboxyl and α-amino groups of consecutive amino acid residues in a polypeptide chain are known as peptide bonds. When the α-amino group of one amino acid combines with the α-carboxyl group of another amino acid, a peptide bond is formed with elimination of a water molecule.
Three types of secondary structures are regularly seen in the proteins:
- α-helix
- β-pleated sheet
- reverse turn or β-bend.
The α-helix is formed by intrachain hydrogen bonds formed between the H atom attached to a peptide N and the carbonyl O of the residue fourth in line behind, in the same polypeptide chain.
Interchain hydrogen bonds are formed between the H atom attached to a peptide N and the carbonyl O of the neighboring polypeptide chains, or segments of a polypeptide chain forming β-pleated sheet structure. There are two types of β-sheets: antiparallel β-sheet, and parallel β-sheet.
In antiparallelβ-pleated sheet the adjacent polypeptide chains run in opposite direction, and the hydrogen bonds are perpendicular to the polypeptide backbone.
In parallelβ-pleated sheet, the adjacent polypeptide chains run in the same direction, and the hydrogen bonds are slant and evenly spaced.
Reverse turn or β-bend is stabilized by hydrogen bonding formed between H atom attached to a peptide N and the carbonyl O of different peptide bonds.8
- Discuss the various forces that stabilize secondary and tertiary structures of proteins. What happens to these forces when protein is denatured?
The secondary and tertiary structure of proteins are mainly stabilized by non-covalent bonds. The non-covalent bonds are:
- Hydrogen bonds
- Hydrophobic interactions
- Electrostatic interactions and
- Van der Waals forces.
- Covalent bonds like disulfide bonds also contribute to the stability of secondary and tertiary structures of proteins. Disulfide bond is formed due to oxidation of –SH groups of two cysteine residues present on the same or different polypeptide chains.
Hydrogen bonds: These are formed when a hydrogen atom is shared between the hydrogen donor atom and the hydrogen acceptor atom. Hydrogen accepting groups are –COO of aspartic acid and glutamic acid, –CO of peptide bond, -O of water, and -N of imidazole. Polar R groups present on the surface of globular proteins primarily form hydrogen bonds with water molecules.
Hydrophobic interactions: These are formed due to coming together of nonpolar (hydrophobic) side chains by eliminating water molecules. Globular structure of the polypeptide chain is due to grouping of nonpolar side chains in the interior portion of the protein molecule, while exposing the polar side chains to the aqueous environment.
Electrostatic interactions or ionic bonds: These are the attractive forces between two opposite charges or repulsion between two similar charges. Electrostatic interactions or salt bonds are formed between non-peptide bond forming –COO− and –+NH3 groups. These groups are generally present in the side chains of acidic and basic amino acids of the protein respectively.
Van der Waals interactions: These are noncovalent attractive and repulsive forces operating between all atoms of a polypeptide chain due to oscillating dipoles. Although very weak, van der Waals forces collectively contribute maximum towards the stability of protein structure. They particularly preserve the nonpolar interior structure of the protein.
During denaturation of proteins, all noncovalent interactions like hydrogen bonds, hydrophobic interactions, electrostatic interactions and van der Waals forces are disrupted with the loss of all higher orders of protein structure except primary structure. Only peptide bonds and disulfide bonds remain intact during denaturation of the proteins.9
- What are fibrous proteins and give examples.
Those proteins that have an axial ratio (ratio of length to breadth) of more than 10 are known as fibrous proteins whereas proteins with axial ratios of less than 10 (generally not greater than 3-4) are known as globular proteins.
Fibrous proteins are elongated or needle shaped and has minimum solubility. Examples are collagen, elastin, keratin and fibrinogen.
- Write briefly on ninhydrin, and biuret reactions.
Ninhydrin reaction: when, an aqueous solution of amino acid with ninhydrin is heated, ninhydrin causes oxidative decarboxylation of α-amino acids with the release of CO2, NH3 and an aldehyde with one carbon less (compared to the original amino acid). The reduced ninhydrin (hydrindantin), along with another molecule of ninhydrin, then reacts with the liberated ammonia to form purple or bright blue complex, which is known as Ruhemann's purple. Proline and hydroxyproline will give yellow color with ninhydrin. Ninhydrin reaction is used for detection as well as estimation of amino acids. The N-terminal (free) amino group of proteins also reacts with ninhydrin to form a blue color (proteins do not give a true color reaction).
Biuret reaction: cupric ions in alkaline medium form a coordination complex with the nitrogens of peptide bonds present in peptides or proteins. The complex is violet or pink in color. The minimum requirement for biuret reaction is a tripeptide (two peptide bonds). Biuret reaction is used for detection as well as estimation of peptides and proteins. A compound called biuret (condensation product of two urea molecules) gives a positive test, hence the name of the test. Ammonium and magnesium ions interfere with biuret reaction.
- Write the methods to identify N-terminal and C-terminal amino acids of the proteins.
Using either Sanger's reagent or Edman's reagent, the N-terminal amino acid of the protein can be identified.
Sanger's reagent is 1-fluoro-2,4-dinitrobenzene (FDNB). It reacts with N-terminal (free) amino group of proteins to form a dinitrophenyl (DNP) derivative (of protein). The protein derivative is then completely hydrolyzed to obtain DNP-amino acid (yellow colored). The DNP-amino acid can be identified by chromatography, which indicates the N-terminal amino acid of the protein.
Edman's reagent is phenyl isothiocyanate. It reacts with N-terminal amino group of the protein to form phenylthiocarbamyl derivative of the protein. On treatment with mild acid, phenylthiohydantoin derivative of N-terminal amino acid is cleaved out from the protein and a new N-terminal (free) amino group of the protein is formed.10
The reaction cycle is repeated again. The phenylthiohydantoin-amino acid released each time can be identified by chromatography. It indicates the N-terminal amino acid. By this method the entire amino acid sequence of the protein can be determined and this method is used in automated sequenator machine.
Using carboxypeptidase, the C-terminal amino acid of the protein can be identified.
Carboxypeptidase is an exopeptidase that acts from the C-terminal of the polypeptide. It specifically hydrolyses and releases the C-terminal amino acid from the protein and in turn, a new C-terminus of the protein is formed. The released C-terminal amino acid can be identified by chromatography.
- Write briefly on denaturation of proteins.
Loss of native three-dimensional structure of protein (including all its secondary structural features) with concomitant loss of biological activity, if any, is called denaturation of proteins.
During denaturation, all the higher orders of protein structure are lost except primary structure. Physical agents like heat, vigorous mechanical shaking and radiation, as well as chemical agents like strong acids and alkalis, organic solvents, urea and salicylates cause denaturation of proteins. During denaturation noncovalent forces like hydrogen bonding, hydrophobic interactions, electrostatic interactions and van der Waals interactions are ruptured but not the covalent bonds like peptide bonds or disulfide bonds. The denatured protein is a random coil and has properties that are different from the native form, such as decreased solubility, increased viscosity and changed isoelectric pH. Denaturation is irreversible, with few exceptions.
Renaturation is reversal of denaturation. Regaining of native three-dimensional (3-D) structure by a denatured protein with simultaneous recovery of biological activity, if any, is known as renaturation of the protein. Very few proteins undergo renaturation. Denatured ribonuclease undergoes complete renaturation with full recovery of activity after the removal of denaturing agents, such as urea and β- mercaptoethanol by dialysis. Renaturation indicates that the primary structure of the protein itself dictates the folding of protein into its native 3-D structure. Renaturation may occur spontaneously or in course of time. Denatured immunoglobulins are renatured after removal of urea.11
- Discuss briefly the primary and tertiary structure of proteins.
Primary structure of protein: Primary structure of protein tells us about the content and sequence of amino acids, and the location of disulfide bonds in a protein, held together by peptide bonds. Primary structure has all the information required for the formation of higher orders of protein structure. By convention, the N-terminal amino acid is always written on the left end of the polypeptide chain and the C-terminal amino acid at the right end of the chain. Mutations of DNA (gene) alter the primary structure of the protein, which in turn alters the protein function.
Tertiary structure of the protein: The native three-dimensional structure of a protein with all its secondary structural features is known as tertiary structure of the protein. In the tertiary structure, nonpolar side chain groups are hidden in the interior of the protein molecule whereas hydrophilic polar groups are exposed to the outside aqueous environment. Tertiary structure is stabilized mainly by noncovalent interactions such as hydrogen bond, hydrophobic interactions, electrostatic interactions and van der Waals interactions. Disulfide bonds also contribute to the stability of tertiary structure. Examples are serum albumin, cytochrome c and myoglobin. Chaperones (heat shock proteins) assist in the formation of tertiary structure of proteins.
- Describe the α-helix of proteins.
The α-helix is a type of secondary structure of the proteins. It is a right-handed helix (spiral structure). The α-helix is formed by intrachain hydrogen bonds formed between the H atom attached to a peptide N and the carbonyl O of the residue fourth in line behind in the same polypeptide chain. The peptide bonds form the backbone of the helix and the side chains of amino acids project outward (from the helix axis). Each turn of the helix contains 3.6 amino acids and pitch of the helix is 0.54 nm (5.4A°). The distance between adjacent amino acids in the chain is 0.15 nm (1.5A°) and the diameter of the helix is 0.6 nm (6A°). Myoglobin has the highest content (80%) of α-helix and chymotrypsin is devoid of α-helix. The α-helix is the most common and stable conformation for a polypeptide chain. Proline is a helix breaking amino acid.12
- Classify proteins with suitable examples.
Based on composition and solubility, proteins are classified into three groups: simple proteins, conjugated proteins, and derived proteins.
- Simple proteins on hydrolysis yield only amino acids. Simple proteins are further classified based on their solubility.
- Albumins are soluble in water, coagulated by heat, and are precipitated by full saturation with ammonium sulfate. Examples are serum albumin, lactalbumin and soya bean albumin.
- Globulins are insoluble in water, but soluble in dilute salt solutions. They are coagulated by heat, and are precipitated by half saturation with ammonium sulfate. Examples are lactoglobulins, serum globulins and egg globulins.
- Glutelins are soluble in dilute acids and alkalies, but insoluble in neutral solvents. Examples are glutelin of wheat and oryzenin of rice.
- Prolamines are soluble in 70% alcohol and are rich in proline. Examples are gliadin of wheat and zein of corn.
- Protamines are small basic proteins. They are soluble in ammonium hydroxide. They are not coagulated by heat. They are nucleoproteins found in sperm, and salmine of Salmon sperm. It is used in the preparation of protamine zinc insulinate.
- Histones are strongly basic proteins. They are soluble in water and dilute acids but insoluble in ammonium hydroxide. Examples are thymus histones and histones of codfish.
- Scleroproteins are structural proteins. They are insoluble in water, salt solutions and organic solvents and soluble only in hot strong acids. Examples are collagen, keratin and elastin.
Conjugate proteins on hydrolysis, yield amino acids and a non-protein part called prosthetic group or conjugated group. Based on the conjugated group present, they are further classified as follows:
- Nucleoproteins are the proteins attached to nucleic acids (DNA or RNA), where nucleic acid is the prosthetic group. Examples are nucleohistones, nucleoprotamines and ribosomes.
- Glycoproteins are the proteins conjugated with carbohydrates. If the carbohydrate content of the molecule is more than 10% it is called mucoprotein. Examples of glycoproteins are immunoglobulins, blood group antigens, and egg albumin.
- Lipoproteins are the proteins attached with lipids, which serve as prosthetic group. Examples are serum lipoproteins, and egg yolk lipoproteins.
- Chromoproteins are the proteins containing colored prosthetic groups. Examples are hemoglobin (red colored), cytochromes (red colored) and visual purple (purple colored).
- Phosphoproteins are the proteins that contain phosphate as prosthetic groups. Examples are casein of milk and vitelline of egg yolk.
- Metalloproteins are the proteins containing metal ions as the prosthetic groups. Examples are carbonic anhydrase (Zn2+), myoglobin (Fe2+) and tyrosinase (Cu2+).
Derived proteins are derived from native proteins. They are of two types: primary derived proteins, and secondary derived proteins.
- Primary derived proteins are obtained by denaturation or coagulation of native proteins. Examples are acid or alkali metaproteins, heat-coagulated proteins, and fibrin formed from fibrinogen.
- Secondary derived proteins are the various products obtained during hydrolysis of the native protein. They are proteoses, peptones, and peptides. Examples are albumose (from albumin) globulose (from globulin) peptone (from fish protein) and oligopeptides.
- Discuss briefly the light absorption properties of amino acids and its application.
Amino acids are colorless because they do not absorb visible light. Aromatic amino acids tryptophan, tyrosine and phenylalanine absorb ultraviolet (UV) light above 240 nm. Aromatic amino acids, particularly tryptophan, absorb high wavelength (250-280 nm) UV light. Most of the UV light absorption of proteins above 240 nm wavelengths is due to tryptophan content. This property of light absorption is most conveniently used for rapid estimation of protein content in solution.